|

 Introduction Introduction
Zinc finger nucleases (ZFNs) are synthetic proteins consisting
of an engineered zinc finger DNA-binding domain fused to the
cleavage domain of the FokI restriction endonuclease. ZFNs
can be used to induce double-stranded breaks (DSBs) in specific
DNA sequences and thereby promote site-specific homologous
recombination and targeted manipulation of genomic loci in
a variety of different cell types. A long-term goal of the
Zinc Finger Consortium is to develop ZFNs as broadly applicable
and readily accessible molecular tools for performing targeted
genetic alterations. The ability to alter the sequence or
structure of any gene of interest would be enormously useful
for biological research and molecular therapeutics.
Highly
efficient, targeted genome manipulation induced by ZFNs
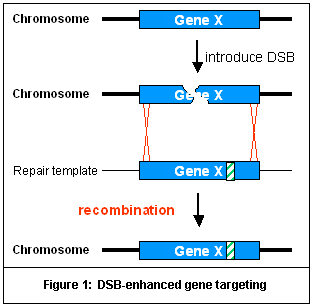
Gene targeting is a method to repair or inactivate any desired
gene of interest. Gene targeting strategies use the introduction
of a double-stranded break (DSB) into a genomic locus to enhance
the efficiency of recombination with an exogenously introduced
homologous DNA "repair template" (Figure 1). DSBs
can stimulate recombination efficiency several thousand-fold,
approaching gene targeting frequencies as high as 50%. ZFNs
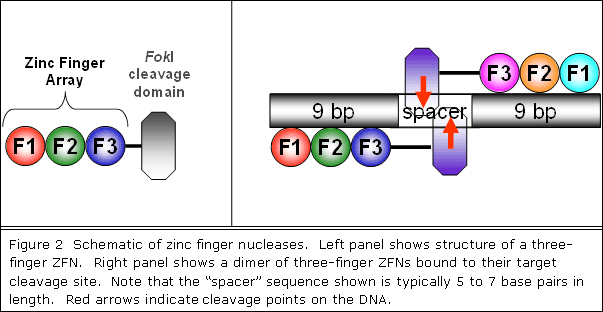
can be used to introduce targeted DSBs. ZFNs consist of a
DNA-binding zinc finger domain (composed of three to six fingers)
covalently linked to the non-specific DNA cleavage domain
of the bacterial FokI restriction endonuclease (Figure 2,
left panel). ZFNs can bind as dimers to their target DNA sites,
with each monomer using its zinc finger domain to recognize
a "half-site" (Figure 2, right panel). Dimerization
of ZFNs is mediated by the FokI cleavage domain which cleaves
within a five or six base pair "spacer" sequence
that separates the two inverted "half sites" (Figure
2, right panel). Importantly, because the DNA-binding specificities
of zinc finger domains can in principle be re-engineered using
one of various methods, customized ZFNs can theoretically
be constructed to target nearly any gene sequence.
Zinc
Finger Engineering
Publicly
available methods for engineering zinc finger domains include:
(1) Context-dependent Assembly (CoDA), (2) Oligomerized Pool
Engineering (OPEN), and (3) Modular Assembly. The Zinc Finger
Consortium provides resources and reagents for practicing
each of these methods (see the Software Tools, Protocols,
Reagents, and ZFC Newsgroup pages).
Members
of the Zinc Finger Consortium continue to work to develop
robust, publicly available methods for engineering zinc finger
nucleases that function well in various cellular environments.
All members have committed to making all protocols, software,
DNA sequences, and reagents they publish available to the
academic scientific community.
|